
Custom Search
Thursday, November 15, 2012
Tuesday, August 28, 2012
PhD in Molecular Medicine / Computational Biology In Italy
PhD in Molecular Medicine / Computational Biology
- DURATION: 4 years
- AWARDING BODIES: University of Milan
- LOCATION: Campus IFOM-IEO Milan
- COORDINATOR OF THE PROGRAM: Pier Giuseppe Pelicci
- SCIENTIFIC COORDINATORS: F. Ciccarelli, A. Ciliberto
- DESCRIPTION:
This course is intended to students having a specific interest for quantitative aspects of biology, such as genomics, systems biology, and modeling of molecular networks. - RESEARCH TOPICS:
Systems Biology, Analysis of Next-Gen Sequencing Data, Network Biology, Evolutionary Genomics, Bioinformatics, Cancer Genomics - SUPERVISION:
Each PhD student will be tutored by three advisors:
The Supervisor is the scientific head of the host laboratory, who will guide the work of the student during his/her PhD period.
The Co-supervisor will contribute in tutoring the student with particular emphasis to the technical and quantitative aspects of the project.
The External Co-supervisor is a foreign expert, who will provide advice at critical stages of the project and meet the student at least once during his/her PhD period. -
TRAINING OFFERED:
Courses: Courses cover the first three years and address basic and advanced topics in cancer biology from a computational perspective.
Seminars: Students are exposed to a wide selection of seminars from international speakers. - FELLOWSHIP:
Tuition and salaries for students are fully covered for the entire PhD period. Refer to "financial matters" for more info. - WORKING LANGUAGE: English
- HOW TO APPLY:
Applications are accepted exclusively online once a year in the period July-September. Go to application process. - STARTING DATE:: November 1st of each year
Thursday, August 2, 2012
Biochemistry - Donald Voet, Judith G. Voet - 4th Edition
Biochemistry is a field of enormous fascination and utility, arising, no doubt, from our own self-interest. Human welfare, particularly its medical and nutritional aspects, has been vastly improved by our rapidly growing understanding of biochemistry. Indeed, scarcely a day passes without the report of a biomedical discovery that benefits a significant portion of humanity. Further advances in this rapidly expanding field of knowledge will no doubt lead to even more spectacular gains in our ability to understand nature and to control our destinies. It is therefore essential that individuals embarking on a career in biomedical sciences be well versed in biochemistry.
Friday, July 13, 2012
7 Tips for Optimal DNA Transfection
- Adapt the transfection conditions to your experiment. Transfection conditions should be optimized according to the cells, the size of the plasmid, the number of plasmids, the expression pattern of the gene of interest, the culture vessel, and the transfection reagent. Reoptimize the conditions for each new cell line.
- Transfect healthy cells. Passage cells at least twice after thawing to allow recovery before transfection, and use cells at low passage number (< 20 passages). Discard overconfluent cells. Regularly check for mycoplasma contaminations. Seed cells the day before transfection accordingly to the confluency recommended by the transfection reagent provider.
- Follow the transfection reagent protocol. Some reagents are inhibited by serum or antibiotics, while others may be used in serum- and antibiotic-containing medium, hence reducing the risks of toxicity and the number of steps in the protocol. Check the recommended cell confluency, as well as the recommended DNA amount and reagent volume.
- Work with high-quality DNA preparation. Check for RNA contamination by agarose gel electrophoresis and ethidium bromide staining. Measure UV absorbance at 280 nm. OD260/280 ratio should reach at least 1.8. Resuspend the plasmid in deionized water or TE buffer at a concentration of ca. 1 µg/µl. Aliquot the plasmid preparation and store at -20°C to avoid freeze/thaw cycles.
- Check serum quality. Some serum lots may inhibit drastically transfection efficiency, hence resulting in lower silencing efficiency. Check transfection efficiency of different serum lots before purchasing a new batch of serum. Also ensure that the medium used allows for efficient transfection, as some media coumpound may decrease transfection efficiency.
- Minimize cytotoxicity by using low DNA amount and low transfection reagent volume. Check that the target gene does not affect cell viability. Analyze transfection at an earlier time point (24 h after transfection instead of 48 h for instance).
- Use appropriate controls. Use a reporter gene to set up and optimize transfection conditions, as those may vary depending on the cells to transfect. Various reporter systems are commercially available: Renilla Luciferase and GFP (Green Fluorescent Protein) are the most commonly used.
DNA transfection
is a main tool for current genetics as well as cell and molecular
biology studies. Understanding the underlying mechanisms and the
different parameters affecting transfection is crucial for optimal,
reproducible, and trustable results. Following these seven tips will
ensure reliable results.
Thursday, July 5, 2012
Wednesday, July 4, 2012
Tuesday, July 3, 2012
Desmoid tumor: benign but nasty

Desmoid tumor, also
called deep-seated fibromatosis, is a benign tumor that is nonetheless a nasty
beast. It is composed of fibroblasts, and it often presents as a large, infiltrative
masses. Desmoid tumors may occur in patients with familial adenomatous
polyposis (also called Gardner syndrome), an autosomal dominant disorder
characterized by innumerable colon polyps (hence the name), as well as other
lesions such as osteomas, jaw osteomas, odontomas, thyroid carcinoma,
epidermoid cysts, fibromas, and sebaceous cysts. If the colon is not removed in
a patient with familial adenomatous polyposis, the chance of colon carcinoma is
virtually 100%.
Back to desmoid
tumors. These tumors usually present as firm, rubbery, ill-defined,
infiltrative masses. They may occur in extra-abdominal regions (for example, in
the shoulder or thigh), within the abdominal wall itself (often attached to the
rectus abdominus), or in the intra-abdominal region. Histologically, desmoid
tumors are composed of bland-appearing fibroblasts in broad fascicles that
infiltrate surrounding tissue. Check out the photo above: you can see muscle
fibers being splayed apart by benign-looking, round-to-spindly tumor cells.
Although benign,
desmoid tumors can act nasty. They are locally aggressive and invasive, and if
you don't excise them completely, they are likely to recur. Treatment with
things that you normally only use for malignancies, such as tamoxifen,
chemotherapy and radiation, has been effective in many cases.
Tuesday, June 12, 2012
Mastocytosis
Mastocytosis is actually a spectrum of rare disorders,
all of which are characterized by - not surprisingly - an increase in mast
cells. Most patients have disease that is localized to the skin, but about
10% of patients have systemic involvement. There is a localized, cutaneous form
of mastocytosis called urticaria pigmentosum that happens mostly in children
and accounts for over half of all cases of mastocytosis.
Clinically, the skin lesions of
mastocytosis vary in appearance. In urticaria pigmentosum, the lesions are
small, round, red-brown plaques and papules. Other cases of mastocytosis show
solitary pink-tan nodules that may be itchy or show blister formation. The
itchiness is due to the release of mast cell granules (which contain histamine
and other vasoactive substances).
In systemic mastocytosis, patients have
skin lesions similar to those of urticaria pigmentosum - but there is also mast
cell infiltration of the bone marrow, lymph nodes, spleen and liver. Patients
often suffer itchiness and flushing triggered by certain foods, temperature
changes, alcohol and certain drugs (like aspirin).
Histologically, mast cell infiltration
may be subtle (with ocasional spindle-shaped mast cells around vessels in the
superficial dermis) to dramatic (with tightly-packed mast cells in the upper to
mid-dermis - check out the photo above). Mast cells may be difficult to
distinguish in regular H&E sections; you may need a metachromatic stain
(toluidine blue or Giemsa) to really visualize the granules.
Tuesday, May 22, 2012
In Virto Fertilization IVF Third Edition - Brian Dale, Kay Elder
Review of the previous edition: '... an excellent handbook for good IVF laboratory practice right down to a description of total quality management (TQM) systems ... concise, well written and should be mandatory reading for all potential and actual embryologists, as well as for those clinicians seeking a wider understanding of the subject.' Human Fertility
Review of the previous edition: 'The authors are masters of this work. Their views are up-to-date, practical, and well documented. This work will be of greatest interest to clinical practitioners of in vitro fertilization that want a relatively short and very precise overview of laboratory methods. It is highly recommended.' John E. Buster, Fertility and Sterility
Book Description
Fully updated with new practical techniques, this new edition describes the form and function of human gametes and embryos, and how this information is used in modern IVF practice. Packed with a wealth of practical and scientific detail, this popular guide is a must for all IVF students and practitioners.
Friday, May 4, 2012
Molecular Cloning: A Laboratory Manual, Volume 1,2,3 by Joseph Sambrook, David William Russell
Book Description
The first two editions of this manual have been mainstays of molecular biology for nearly twenty years, with an unrivalled reputation for reliability, accuracy, and clarity.
In this new edition, authors Joe Sambrook and David Russell have completely updated the book, revising every protocol and adding a mass of new material, to broaden its scope and maintain its unbeatable value for studies in genetics, molecular cell biology, developmental biology, microbiology, neuroscience, and immunology.
Handsomely redesigned and presented in new bindings of proven durability, this three-volume work is essential for everyone using today’s biomolecular techniques.
The opening chapters describe essential techniques, some well-established, some new, that are used every day in the best laboratories for isolating, analyzing and cloning DNA molecules, both large and small.
These are followed by chapters on cDNA cloning and exon trapping, amplification of DNA, generation and use of nucleic acid probes, mutagenesis, and DNA sequencing.
The concluding chapters deal with methods to screen expression libraries, express cloned genes in both prokaryotes and eukaryotic cells, analyze transcripts and proteins, and detect protein-protein interactions.
The Appendix is a compendium of reagents, vectors, media, technical suppliers, kits, electronic resources and other essential information.
As in earlier editions, this is the only manual that explains how to achieve success in cloning and provides a wealth of information about why techniques work, how they were first developed, and how they have evolved.
Book Info
Peter MacCallum Cancer Institute, Melbourne, Australia. Updated text includes revised protocols and new material for students in genetics, molecular cell biology, developmental biology, microbiology, neuroscience, and immunology. Describes techniques, cDNA cloning and more. Softcover, hardcover also available. DNLM: Cloning, Molecular--Laboratory Manuals.
The first two editions of this manual have been mainstays of molecular biology for nearly twenty years, with an unrivalled reputation for reliability, accuracy, and clarity.
In this new edition, authors Joe Sambrook and David Russell have completely updated the book, revising every protocol and adding a mass of new material, to broaden its scope and maintain its unbeatable value for studies in genetics, molecular cell biology, developmental biology, microbiology, neuroscience, and immunology.
Handsomely redesigned and presented in new bindings of proven durability, this three-volume work is essential for everyone using today’s biomolecular techniques.
The opening chapters describe essential techniques, some well-established, some new, that are used every day in the best laboratories for isolating, analyzing and cloning DNA molecules, both large and small.
These are followed by chapters on cDNA cloning and exon trapping, amplification of DNA, generation and use of nucleic acid probes, mutagenesis, and DNA sequencing.
The concluding chapters deal with methods to screen expression libraries, express cloned genes in both prokaryotes and eukaryotic cells, analyze transcripts and proteins, and detect protein-protein interactions.
The Appendix is a compendium of reagents, vectors, media, technical suppliers, kits, electronic resources and other essential information.
As in earlier editions, this is the only manual that explains how to achieve success in cloning and provides a wealth of information about why techniques work, how they were first developed, and how they have evolved.
Book Info
Peter MacCallum Cancer Institute, Melbourne, Australia. Updated text includes revised protocols and new material for students in genetics, molecular cell biology, developmental biology, microbiology, neuroscience, and immunology. Describes techniques, cDNA cloning and more. Softcover, hardcover also available. DNLM: Cloning, Molecular--Laboratory Manuals.
Friday, April 27, 2012
Aplastic anemia
Aplastic anemia falls into the category of "anemias-in-which-the-cells-
In aplastic anemia, however, there's a giant clue staring you straight in the face: pancytopenia. Not only are the red cells decreased in number, but so are the white cells and platelets. That's because the marrow is basically empty. Check out the image above of a bone marrow biopsy from a patient with aplastic anemia. There's supposed to be hematopoietic tissue in there - but basically all you see are mature red cells and lymphocytes. All precursors are decreased: erythroblasts, granulocyte precursors, and megakaryocytes. So when you look at the blood, the mature cells in the blood are all decreased too.
Clinically, patients present with just what you'd expect: findings related to their cytopenias. Patients will usually be pale and fatigued (from the anemia), with bleeding and bruising (from the thrombocytopenia) and recurrent infections (from the leukopenia). Lots of things can cause aplastic anemia (drugs, viruses, pregnancy, Fanconi anemia), but many times, no cause is found.
Patients are usually treated with blood products as needed, and if the aplasia doesn't resolve, then drugs such as G-CSF and prednisone are given. Bone marrow transplant is successful in many patients, but because of the high morbidity and mortality of the procedure, it's used only as a last resort. For a serious hematologic disorder, the prognosis is relatively good: 3 year survival is on the order of 70%.
Monday, April 23, 2012
Trouble-shooting: Non-specific bands
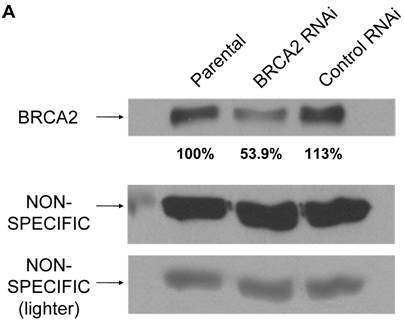
Here are some troubleshooting hints that we have gathered regarding non-specific bands after PCR reactions:
Non-optimal Mg++ concentration
Nucleotide concentration is too high or unbalanced
DNA contamination / carryover
Primer annealing temperature is too low
Mispriming caused by secondary structure of template, snapback, or excessive homology at 3' ends of primers
Primers are degraded or sequence is not optimal
Primer concentration is too high
Cycle number is too high
Incorrect template to enzyme ratio
Non-optimal Mg++ concentration
Suggestion: Titrate magnesium concentration using our PCR Optimization Kit.
Nucleotide concentration is too high or unbalanced
The standard concentration is 20-200 µM of each nucleotide Suggestions:
Check the concentration of stock solutions of all nucleotides
Double check the final concentrations of all nucleotides
DNA contamination / carry-over
Suggestions:
Test for carryover by performing PCR without adding target DNA.
Avoid carryover
To prevent carryover, good lab practices should be used such as:
Physically isolating PCR preps and products
Autoclaving solutions, tips, and tubes
Aliquoting reagents to minimize repeated sampling (no more than 20 reactions per aliquote)
Eliminating aerosols by using positive displacement pipettes
Premixing reagents
Adding DNA to reaction last
Choosing (+) and (-) controls carefully
Soaking gel box and combs in 1M HCl to depurinate DNA
Using new razor blades to exise bands
Using new razor blades to exise bands
Covering UV box with fresh plastic wrap
Always using oil overlay
To eliminate contamination / carryover:
UV irradiation: Mix all components, except template DNA, irradiate in clear 0.5 ml polypropylene tubes in direct contact with glass transilluminator (254 and 300 nm UV bulbs) for 5 minutes
UNG Digestion: Incorporate d-UTP nucleotides into reaction and do subsequent uracyl DNA glycosylase digestion
Primer annealing temperature is too low
Primer annealing temperature is typically 50-60°C (may be higher or lower based on primer sequence and buffer components).
Suggestion: Determine Tm/annealing temperature based on one of the following equations:
If primers are 20-35 bases If primers are 14 - 70 bases
Tp = 22 + 1.46(Ln)
Ln = 2(# G or C) + (# A or T)
TP = Effective annealing
temperature ± 2 - 5 Tm = 81.5 + 16.6 (log10 [J+]) + 0.41
(% G + C) - (600/l) - 0.063
(% Formamide) + 3 to 12
[J+] = concentration of Monovalent cations
l = length of oligo
Mispriming caused by secondary structure of template, snapback, or excessive homology at 3' ends of primers
Suggestions:
Increase initial denaturation temperature to 95-97°C
Denature DNA minus enzyme & buffer for 4-6 minutes
Increase cycling denaturation time 15-30 seconds
Try "Hot Start" technique
Add T4 Gene 32 protein 3-5 µl/ml
When designing primers, make sure there is no more than 2 bases of homology at the 3' end. Use a primer design program if available
Consider addition of cosolvent to reaction buffer:
3-15% DMSO
1-10% Formamide
5-15% Polyethylene glycol
10-15% glycerol
Primers are degraded or sequence is not optimal
Primers should have same number A & T's versus G & C's, and they should be at least 14 bases for specificity. Suggestions:
If primers are short and A-T rich, add 0.9 - 2.0%(v/v) DMSO
If primers are G-C rich, add 1-10% (v/v) Formamide
Double check priming sequence, use primer design program if available
Check aliquot of primers on a gel to ensure they are not degraded
Primer concentration is too high
Suggestion: Adjust the primer concentration (0.1 - 1.0 µM of each primer is optimal)
Cycle number is too high
Most templates require 25-30 cycles. Suggestion: Cycle number should be based on starting concentration of template DNA.
If the number of target molecules in your sample is...
Then we recommend the following number of Cycles...
3 x 105 25-30
1.5 x 104 30-35
1.0 x 103 35-40
50 40-45
Incorrect template to enzyme ratio
The necessary amount of template varies from reaction to reaction. As a guideline, 100 - 750 ng human DNA (105 - 106 copies) per 100 µl reaction. The amount of enzyme should be optimized for each template. Suggestions:
Titrate the amount of template in the reaction
Perform optimization experiment varying enzyme concentration by 0.50 increments in suggested range (0.5 to 5.0 units)
Monday, April 9, 2012
Foundations in microbiology by Talaro 8th Edition
Talaro/Chess: Foundations in Microbiology is an allied health microbiology text for non-science majors with a taxonomic approach to the disease chapters. It offers an engaging and accessible writing style through the use of tools such as case studies and analogies to thoroughly explain difficult microbiology concepts.
We are so excited to offer a robust learning program with student-focused learning activities, allowing the student to manage their learning while you easily manage their assessment. Detailed reports show how your assignments measure various learning objectives from the book (or input your own!), levels of Bloom’s Taxonomy or other categories, and how your students are doing.
The Talaro Learning program will save you time while improving your students success in this course.
We are so excited to offer a robust learning program with student-focused learning activities, allowing the student to manage their learning while you easily manage their assessment. Detailed reports show how your assignments measure various learning objectives from the book (or input your own!), levels of Bloom’s Taxonomy or other categories, and how your students are doing.
The Talaro Learning program will save you time while improving your students success in this course.
Sunday, April 8, 2012
Cystic Fibrosis
Big Advance Against Cystic Fibrosis: Stem Cell Researchers Create Lung Surface Tissue in a Dish
Harvard stem cell researchers at Massachusetts General Hospital (MGH) have taken a critical step in making possible the discovery in the relatively near future of a drug to control cystic fibrosis (CF), a fatal lung disease that claims about 500 lives each year, with 1,000 new cases diagnosed annually.
Beginning with the skin cells of patients with CF, Jayaraj Rajagopal, MD, and colleagues first created induced pluripotent stem (iPS) cells, and then used those cells to create human disease-specific functioning lung epithelium, the tissue that lines the airways and is the site of the most lethal aspect of CF, where the genes cause irreversible lung disease and inexorable respiratory failure.
That tissue, which researchers now can grow in unlimited quantities in the laboratory, contains the delta-508 mutation, the gene responsible for about 70 percent of all CF cases and 90 percent of the ones in the United States. The tissue also contains the G551D mutation, a gene that is involved in about 2 percent of CF cases and the one cause of the disease for which there is now a drug.
The work is featured on the cover of this month's Cell Stem Cell journal. Postdoctoral fellow Hongmei Mou, PhD, is first author on the paper, and Rajagopal is the senior author.
Mou credits learning the underlying developmental biology in mice as the key to making tremendous progress in only two years. "I was able to apply these lessons to the iPS cell systems," she said. "I was pleasantly surprised the research went so fast, and it makes me excited to think important things are within reach. It opens up the door to identifying new small molecules [drugs] to treat lung disease."
Doug Melton, PhD, co-director of the Harvard Stem Cell Institute, said, "This work makes it possible to produce millions of cells for drug screening, and for the first time human patients' cells can be used as the target." Melton, who is also co-chair of Harvard's inter-School Department of Stem Cell and Regenerative Biology and is the Xander University Professor, added, "I would expect to see rapid progress in this area now that human cells, the very cells that are defective in the disease, can be used for screening."
Rajagopal said, "The key to our success was the ecosystem of the Harvard Stem Cell Institute and MGH. HSCI investigators pioneered the strategies we used, helped us at the bench, and gave us advice on how to combine our knowledge of lung development with their exciting new platforms. Indeed, we also enjoyed a wonderful collaboration with Darrell Kotton's lab at Boston University that was able to convert mouse cells into lung tissue. These interactions really helped fuel us ahead."
The epithelial tissue created by Rajagopal and his colleagues at the MGH Center for Regenerative Medicine also provides researchers with the same cells that are involved in a number of common lung conditions, including asthma, lung cancer, and chronic bronchitis, and may hasten the development of new insights and treatments into those conditions as well.
"We're not talking about a cure for CF; we're talking about a drug that hits the major problem in the disease. This is the enabling technology that will allow that to happen in a matter of years," said Rajagopal, a Harvard Medical School assistant professor of Medicine.
Also a physician trained as a pulmonologist, the specialty that treats CF patients, Rajagopal said, "When we talk about research and advances, donors and patients ask: 'When? How soon?' And we usually hesitate to answer. But we now have every single piece we need for the final push. So I have every hope that we'll have a therapy in a matter of years."
Cystic fibrosis, which used to claim its victims in infancy or early childhood, has evolved into a killer of those in their 30s because treatments of the infections that characterize the disease have improved. But despite those advances, there has been little progress in treating the underlying condition that affects the vast majority of patients: a defect in a single gene that interferes with the fluid balance in the surface layers of the airways and leads to a thickening of mucus, difficulty breathing and repeated infections and hospitalizations.
The discovery and recent FDA approval of the drug Ivacaftor, which corrects the G551D defect seen in about 2 percent of CF patients, has served as a proof of concept to demonstrate that the disease can be attacked with a conventional molecular treatment. In fact, Ivacaftor was found by screening thousands of drugs on a far less than ideal cell line. In the end, many drugs that functioned well on this cell line proved ineffective when used on genuine human airway tissue.
Genuine human airway tissue is the gold standard prior to drugs being tested clinically, but it has been extremely difficult to obtain the tissue from patients, and when it could be obtained, the tissue rarely survived long in the lab -- all of which created a major bottleneck in screening for a therapy. But by creating iPS cells that contain the entire genome of a CF patient and directing those cells to develop into lung progenitor cells, which then develop into epithelium, the group appears to have solved this key problem.
Rajagopal, who did his own postdoctoral fellowship in Melton's laboratory during the first half of the past decade after completing his training in pulmonary medicine, said that having both the G551D and 508 genes in the epithelial tissue provides a way to prove that the tissue will be effective in testing drugs against CF.
"We've created the perfect cell line to show that the drug out there that works against G551D mutation works in this system, and then we're in business to screen for a drug against delta 508," he said. "We'll know soon that the cell line works. We know it makes bonafide airway epithelium, and we'll have the proof of principle that the tissue responds properly to the only known drug. We think this is the near-ideal tissue platform to find a drug for the majority of CF."
Rajagopal's lab has created numerous other cell lines to further show that a CF drug that works in one patient should work in others and to see whether this will be an area that allows a more personalized approach to medicine.
Saturday, March 24, 2012
Biochemistry by Jeremy M Berg, John L Tymoczko, Lubert Stryer 6th Edition
One of the most trusted books in the field, Biochemistry still shapes the way biochemistry is taught. Clear and concise in their writing and illustration, Berg et al. emphasize physiological and medical relevance. This edition includes new coverage on gene regulation, new examples from medicine and the latest experimental techniques
Tuesday, March 20, 2012
Griffiths - An Introduction to Genetic Analysis (9th Edition)
The author team welcomes a new coauthor, Sean B. Carroll, a recognized leader in the field of evolutionary development, to this new edition of Introduction to Genetic Analysis (IGA). The authors' ambitious new plans for this edition focus on showing how genetics is practiced today. In particular, the new edition renews its emphasis on how genetic analysis can be a powerful tool for answering biological questions of all types.
Subscribe to:
Comments (Atom)








