UniSeq™ is a universal DNA sequencing primer walking technology developed by Nucleics for large scale DNA sequencing applications like whole genome sequencing projects. The UniSeq DNA sequencing library & software offers the following advantages over other sanger DNA sequencing methodologies:
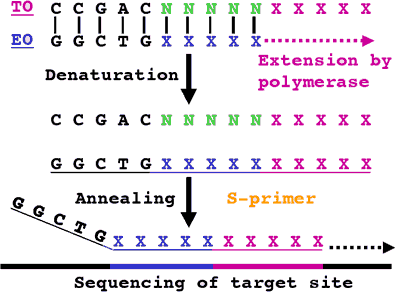
Figure 1. Generation of template specific UniSeq S-primer. The EO hybridizes to the TO and gets extended to form the S-primer. N: degenerated oligonucleotide positions. X: specific positions of variable nucleotide value.
By selecting specific EO and TO oligonucleotides from a small, pre-synthesised library of 768 oligonucleotides, over 131,000 different, template specific primers can be created. This simple combinatorial effect forms the basis for the high specificity and universal applicability of the UniSeq system in DNA sequencing.
The UniSeq system is fully compatible with the common DNA sequencing reagents (e.g. BigDye™ from Applied Biosystems or DYEnamic™ from Amersham Biosciences) and gives excellent results using modern capillary DNA sequencers (Figure 2).
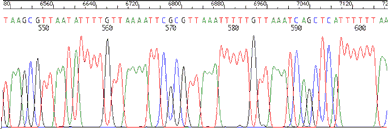
Figure 2. UniSeq reaction sequencing trace. Two hundred nanograms of plasmid DNA was sequenced with BigDye vers.3.1 and UniSeq primers EO 18 and TO 447. The DNA sequencing reaction was purified by ethanol precipitation and analyzed on an ABI 3700 DNA sequencer.
The UniSelect™ DNA sequencing software is provided for the automated and optimized selection of the optimal EO & TO primer pairs for each DNA template. The UniSelect software also supplies an automated interface to control the robotic pipetting of the UniSeq oligonucleotides, DNA templates and other required sequencing reagents.
To aid the finishing of Whole Genome Sequencing (WGS) projects, an additional software package (UniFinish™) is available. UniFinish is able to parse ACE format assembly files and select the EO/TO oligonucleotides, together with the appropriate DNA templates, required to close non-physical DNA sequence gaps in the genome assembly.
UniSeq provides the advantages of both WGS strategies. It offers the speed and simplicity of the RSS approach, while providing the data efficiency inherent with the PWS approach. Computer simulations and limited trials have shown that UniSeq DNA sequencing system offers cost and time savings of greater than 80% over current WGS approaches.
- A fast and cost effective universal primer walking approach
- Very simple and robust protocol
- Compatible with all Sanger DNA sequencing technologies and equipment
- Fully adaptable to high throughput, large scale DNA sequencing facilities
The UniSeq DNA sequencing system
The UniSeq DNA sequencing system utilizes a unique methodology to reliably create template specific DNA sequencing primers. The process of generating each UniSeq DNA sequencing primer involves the addition of an "E"- and "T"-oligonucleotide, together with a specifically formulated additive mixture, directly to the sequencing reaction. These oligonucleotides (termed EO and TO) hybridize during the sequencing reaction to produce the template specific primer (S-primer) (Figure 1).
Figure 1. Generation of template specific UniSeq S-primer. The EO hybridizes to the TO and gets extended to form the S-primer. N: degenerated oligonucleotide positions. X: specific positions of variable nucleotide value.
By selecting specific EO and TO oligonucleotides from a small, pre-synthesised library of 768 oligonucleotides, over 131,000 different, template specific primers can be created. This simple combinatorial effect forms the basis for the high specificity and universal applicability of the UniSeq system in DNA sequencing.
The UniSeq system is fully compatible with the common DNA sequencing reagents (e.g. BigDye™ from Applied Biosystems or DYEnamic™ from Amersham Biosciences) and gives excellent results using modern capillary DNA sequencers (Figure 2).

Figure 2. UniSeq reaction sequencing trace. Two hundred nanograms of plasmid DNA was sequenced with BigDye vers.3.1 and UniSeq primers EO 18 and TO 447. The DNA sequencing reaction was purified by ethanol precipitation and analyzed on an ABI 3700 DNA sequencer.
The UniSelect™ DNA sequencing software is provided for the automated and optimized selection of the optimal EO & TO primer pairs for each DNA template. The UniSelect software also supplies an automated interface to control the robotic pipetting of the UniSeq oligonucleotides, DNA templates and other required sequencing reagents.
To aid the finishing of Whole Genome Sequencing (WGS) projects, an additional software package (UniFinish™) is available. UniFinish is able to parse ACE format assembly files and select the EO/TO oligonucleotides, together with the appropriate DNA templates, required to close non-physical DNA sequence gaps in the genome assembly.
Feature and benefits of UniSeq
Genomes are finished faster
The UniSeq DNA sequencing system provides a competitive alternative to DNA sequencing with custom synthesized oligonucleotides. While many advances have been made in the automation of oligonucleotide synthesis, this process is still complex (eg. requiring high maintenance machinery) and slow (several hours per synthesis). These limitations have become extremely critical for high throughput DNA sequencing facilities where modern capillary DNA sequencers have reduced the separation time to one to two hours. The generation of specific DNA sequencing primers by the UniSeq process during the DNA sequencing reaction step (i.e. no time-costs) breaks the current bottleneck imposed by custom oligonucleotide synthesis and allows for the most efficient utilization of existing DNA sequencing instruments and other equipment.Lower per finished base costs
Currently, two major strategies are used in WGS – Random Shotgun DNA Sequencing (RSS) and Primer Walking DNA Sequencing (PWS). Most WGS projects currently employ RSS, especially in early stages of projects. However, RSS requires generating a large amount of redundant DNA sequence (commonly 6 to 15 times the genome size) for assembly purposes. The alternative PWS strategy requires relatively little redundant DNA sequence data, however, it requires a large numbers of custom made oligonucleotide primers. The high costs (US$3 to 5 per primer), together with the required synthesis delay, have prevented the general adoption of PWS strategy in WGS Genomics projects.UniSeq provides the advantages of both WGS strategies. It offers the speed and simplicity of the RSS approach, while providing the data efficiency inherent with the PWS approach. Computer simulations and limited trials have shown that UniSeq DNA sequencing system offers cost and time savings of greater than 80% over current WGS approaches.


specialize in amino acid analysis and oligo synthesis
ReplyDelete